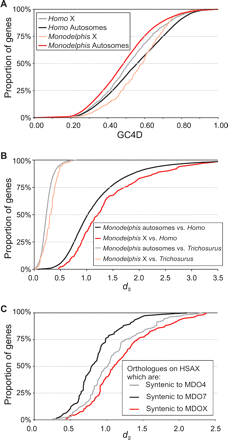
G+C Content at 4D sites (GC4D) and dS in the Monodelphis and Homo X chromosomes. (A) GC4D values of Monodelphis genes in 1:1 orthology relationships with Homo genes. The cumulative distribution of GC4D for orthologs on the Monodelphis X chromosome is shifted to higher values compared with that for orthologs on Monodelphis autosomes, and genes on the Monodelphis X chromosome have a much increased median GC4D. This is exactly the opposite of the situation among Homo orthologs, where X chromosome genes have reduced GC4D. (B) The increased GC4D on the Monodelphis X chromosome is accompanied by an increase in median dS and a rightward shift of the dS cumulative distribution. This effect is even more pronounced when comparing orthologs between Monodelphis and Trichosurus, suggesting that much of the increase in dS has taken place along the marsupial lineage. (C) The dS for Homo–Monodelphis orthologs that lie on the Homo X chromosome and that are syntenic to the Monodelphis chromosome X or to chromosomes 4 and 7. Among orthologs on the Homo X chromosome, only those that are also found on the Monodelphis X chromosome have elevated dS, confirming that this is largely a marsupial-specific phenomenon.











