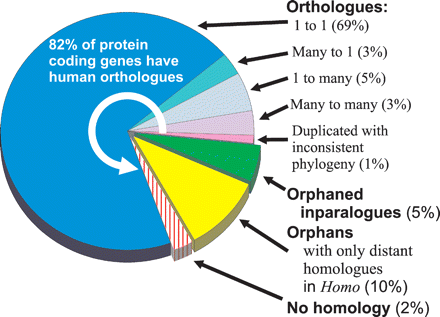
Proportion of Monodelphis domestica genes with Homo orthologs. The majority (82%) of protein coding genes have PhyOP-predicted orthologs among human genes. The classes of orthology relationships are indicated by counts of Homo orthologs followed by Monodelphis. Thus, a one-to-many relationship refers to a single Homo gene that is orthologous to several Monodelphis-specific duplications. Many of the genes with no predicted orthology with a Homo gene are nevertheless closely related to one or more Monodelphis genes with a low dS, indicating that these are Monodelphis-specific duplications. These are labeled as “Orphaned inparalogues.” This class includes genuine biological losses in the eutherian or human lineage, as well as heuristic failures in the PhyOP-prediction pipeline. The latter are often related to errors in gene predictions in difficult cases where there are many adjacent tandem duplications, such as among KRAB-ZnF genes. Similarly, many of the Monodelphis genes which have no plausible candidate ortholog in the human genome (labeled “Orphans with only distant homologues in Homo”) are likely to involve problematic gene predictions. The Monodelphis genes in the “No homology” class do not have significant BLAST alignments (E < 10−5) covering the majority of the sequence (75% of the shorter of the pair of sequences, and involving more than 50 residues). Most of these appear to be fragmentary, chimeric, or erroneous gene predictions, or noncoding sequence.











