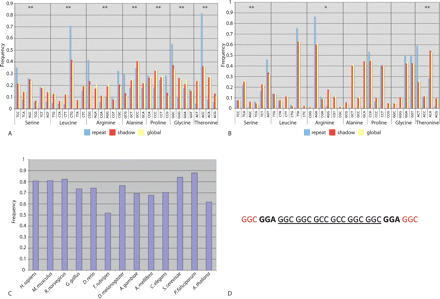
(A,B) Codon usage within a homopeptide (repeat), outside the homopeptide (shadow), and from the codon usage database (global). (A) H. sapiens, (B) P. falciparum. Significant differences between the codon usage within a homopeptide compared with the global codon usage: (*) P ≤ 0.01, (**) P ≤ 0.001. (C) The frequency of codons within a homopeptide that are immediately upstream and downstream of the 5′ and 3′ codons (the capping codons), respectively, which surround a homocodon and are synonymous to the homocodon. (D) An example of the encoding codons of a homopeptide, highlighting the codons mentioned in C. (Underlined) The homocodon, (red) the 5′ and 3′ capping codons, (bold) the immediate upstream and downstream codons of the capping codons.











