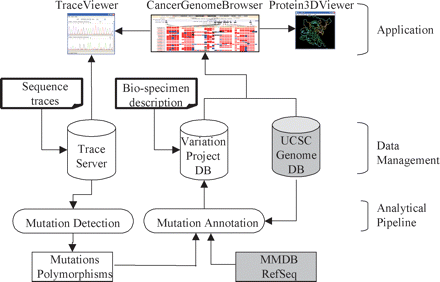
System overview of Cancer Genome WorkBench (CGWB). CGWB takes two sets of input data, sequence traces and biospecimen descriptions, which are loaded into the trace server and the variation database, respectively. The mutation analysis pipeline, which consists of SNPdetector and IndelDetector, processes the traces to detect mutations and polymorphisms. These variations are integrated into the reference human genome and their annotations on protein coding and 3D structure are computed using our HapScope pipeline (Zhang et al. 2002). The mRNA and protein annotation are based on the latest version of NCBI RefSeq database (Wheeler et al. 2006). The 3D structure mapping is based on NCBI’s MMDB database (Wheeler et al. 2006). The variation data are then loaded into the variation database. Application programs then access the databases to generate visual displays.











