N50 length of haplotype segments by varying sequence coverage and polymorphism rate
Click on table to view larger version.
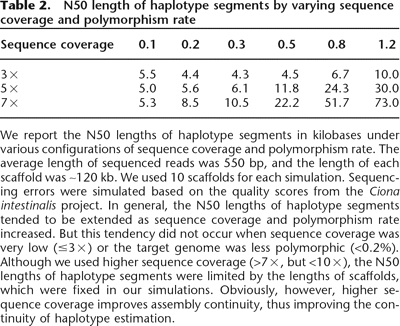
We report the N50 lengths of haplotype segments in kilobases under various configurations of sequence coverage and polymorphism rate. The average length of sequenced reads was 550 bp, and the length of each scaffold was ∼120 kb. We used 10 scaffolds for each simulation. Sequencing errors were simulated based on the quality scores from the Ciona intestinalis project. In general, the N50 lengths of haplotype segments tended to be extended as sequence coverage and polymorphism rate increased. But this tendency did not occur when sequence coverage was very low (≤3×) or the target genome was less polymorphic (<0.2%). Although we used higher sequence coverage (>7×, but <10×), the N50 lengths of haplotype segments were limited by the lengths of scaffolds, which were fixed in our simulations. Obviously, however, higher sequence coverage improves assembly continuity, thus improving the continuity of haplotype estimation.











