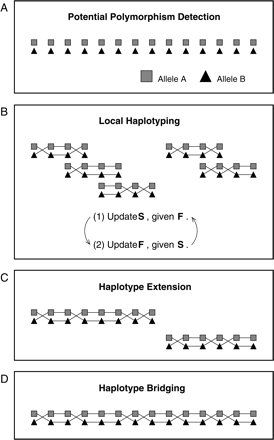
Overview of our haplotype reconstruction strategy. (A) In this step, Bayesian model selection is used to detect potential polymorphic sites along a genome. Two different alleles at each polymorphic site are represented by a triangle and a rectangle. In this figure, detected polymorphic sites run from the left-hand side to the right-hand side according to their genomic positions in an assembly. (B) We determine phases among detected polymorphisms by using a Gibbs sampling method. We focus on estimating reliable haplotypes from a small number of polymorphisms by repeating the following two steps: (1) update S based on F, (2) update F based on S. (C) If adjacent short haplotypes overlap and show a consistency, those haplotypes are combined. (D) Adjacent haplotypes are connected if the fragments spanning those adjacent haplotypes significantly support the connection.











