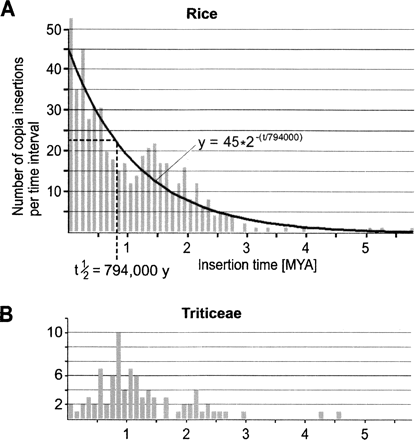
Distribution of copia element insertion times. Estimated insertion times were divided into bins of 100,000 yr. (A) The distribution of rice copia elements can be described as a hyperbolic function based on the assumption that retrotransposon sequences are at least partially removed from the genome at a constant rate. The curve represents the best fit for the starting value (43.6) and the half-life (796,000). (B) Insertion distribution of 87 copia elements from Triticeae (wheat and barley). The distribution is not similar to a hyperbolic distribution.











