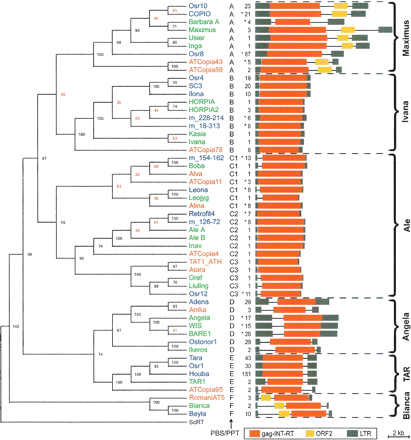
Phylogenetic tree of 52 copia families from Triticeae, rice, and Arabidopsis. Names of copia families from Arabidopsis are printed in red, those from rice in blue, and those from Triticeae in green. Subfamilies are indicated by a capital letter at the end of the name. Black uppercase letters refer to the type of primer binding site (PBS) and polypurine tract (PPT) detailed in Figure 2. Asterisks indicate the presence of additional, closely related families that had been omitted due to space constraints. Copy numbers in these cases refer to the total of all represented families (see Supplemental Fig. 1). Bootstrap numbers at the forks indicate how many times the sequences to the right of the fork occurred in the same group of 100 trees. Strong bootstrap values of at least 80 are shown in black. The copy numbers and sequence organization of all families are displayed next to the respective names. Major evolutionary lineages are indicated by curly brackets. A reverse transcriptase sequence from yeast (ScRT) served as outgroup.











