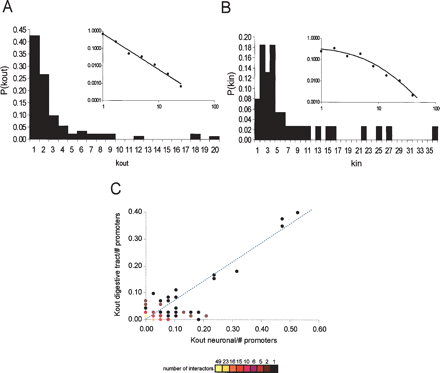
Architecture of the C. elegans core neuronal PDI network. (A) Out-degree distribution. Logarithmic binning was applied to fit a power law to P(kout): P(kout) = 0.69k−2.05 (R2 = 0.99; inset). (B) In-degree distribution. Logarithmic binning was applied to fit a power law with saturation to P(kin) = 1093.2*(10.83 + k)−3.30(R2 = 0.97; inset). (C) Interactor hubs in the core neuronal PDI network have a similar out-degree in the digestive tract PDI network (diagonal). Interactors specific for each network are localized on the axes. The colors indicate the number of interactors per feature.











