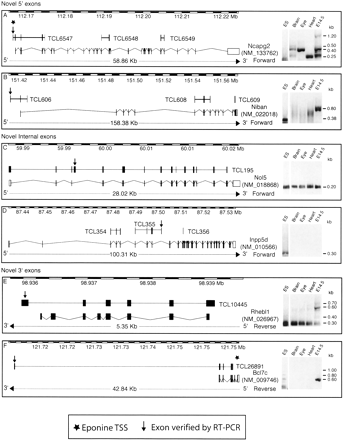
Discovery of novel exons on known RefSeq genes. The figure shows six examples of RefSeq genes to which novel exons (indicated by the arrow) were added using gene trap data, confirmed by RT-PCRs conducted on ES cell RNA as well as four other RNA samples shown in the panel on the right of each diagram. TCL6547 was verified as an alternative 5′-UTR exon of the Ncapg2 gene found to be expressed in all RNA samples tested, showing several splicing variants. The TCL606 cluster also confirms a new 5′-UTR exon (belonging to the Niban gene); however, its expression was only confirmed in ES cells and whole embryo (and not in heart, brain, or eye). The TCL195 cluster represents a novel alternatively spliced “cassette” exon added between exon 3 and exon 4 of the Nol5 gene, which is found to be expressed in all samples tested, always yielding the same PCR product. TCL355 adds an internal exon to the Inpp5d gene, and its sequence terminates at this exon. This cluster was found to be expressed only within ES cells. The TCL10445 cluster adds a 3′-exon to the Rhebl1 gene, and the transcript that includes this exon skips the last two constitutive exons of this gene in all RNA samples tested, while the isoform that includes the exons between is only found in whole embryo RNA. TCL26891 adds a further 3′-exon >30 kb away from the last exon of the Bcl7c gene. This exon is only found expressed in ES-cell RNA and whole embryo RNA.











