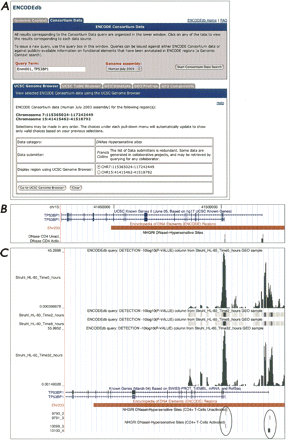
The UCSC Genome Browser query form. (A) A query using “ENm001, TP53BP1” as the search term, searching against the Human July 2003 sequence assembly. Users can select desired data categories and data submitters from the pull-down menus provided on the form; here, DNase hypersensitive sites has been chosen as the data category, and Francis Collins has been chosen as the data submitter. The choices under each pull-down menu will dynamically update to show only valid choices based on any previous selections; this feature was built into ENCODEdb to assist the user, since there is no similar functionality available through the GEO Web site itself. Because the query was performed using two search terms, users have the option to go to the region of ENm001 (on chromosome 7) or TP53BP1 (on chromosome 15). Once the Go To UCSC Genome Browser button is pressed, a new window is spawned, showing the selected experiments within the context of the UCSC Genome Browser (B). Here, the image is centered on two forms of TP53BP1 in the UCSC Known Genes tracks. The Known Genes track was opened manually, and the window was shifted to the right for better viewing of the 5′ ends of the transcripts. Through this type of view, the overlap between known genes and the DNase I hypersensitivity data requested through ENCODEdb becomes obvious. (C) ENCODEdb can be used to test specific hypotheses. This panel illustrates the results of a query issued to determine whether an alternative promoter corresponds to a downstream hypersensitive site. The view was generated by clicking the GEO Components tab shown in Figure 2A and then changing the selections to the following: ChIP-on-chip assay, experimental group “Pol2 ChIP-chip of RA-stimulated HL60 cells at four timepoints” in the HL60 cell line, for the region Chromosome 15:41486698–41590028. Wiggle was chosen as the output format, and the data column to display in this format is Detection. The resulting UCSC Genome Browser view shows the annotation tracks seen in Figure 2B, along with the newly requested ChIP-chip data. The 0- and 32-h timepoint tracks have been manually expanded to illustrate the correlation between hypersensitive sites, the 5′ ends of the genes, and the presence of PolII.











