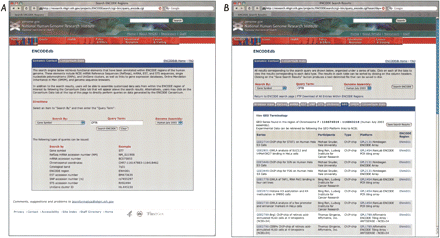
Genomic Context searches using ENCODEdb. (A) Queries can be issued on any genomic region of interest, without knowing in advance whether the region actually lies in an ENCODE region or which ENCODE region it lies in. Searches may be issued using a gene symbol, RefSeq mRNA accession number (NM number), mRNA accession number, chromosomal coordinates, cytological band, ENCODE region, EST accession number, SNP accession number, STS accession number, or UniGene cluster ID; examples of proper syntax for each search is shown in the figure. At present, users may select either the Human July 2003 (hg16) or May 2004 (hg17) assemblies for their searches. (B) The result of a gene symbol-based search, using CFTR as the query. The GEO tab has been highlighted. All GEO Series (GSE) in the CFTR region are shown. Clicking on any of the GSE numbers will spawn a new window, showing the GEO series entry page within GEO. Clicking on any of the GEO platform (GPL) numbers takes the user to the appropriate GEO platform page, giving full information on the array platform used in this experiment. Clicking on ENCODE region numbers takes the user to the appropriate view within the UCSC Genome Browser. A “GEO Terminology” link is provided for users who are unfamiliar with the structure or content of the GEO database.











