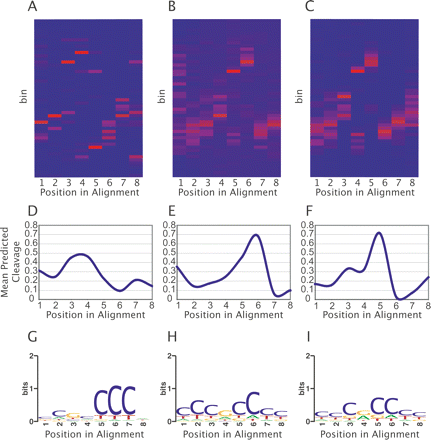
Analysis of representative high-scoring CORCS. (A–C) Heat maps of CORCS found in (A) the MPSS data set and (B) the Union data set using the discrete sampler, and in (C) the Union data set using the continuous sampler. The X-axis represents each position in the CORCS and the Y-axis represents cleavage value bins. Dark blue cells in the heat map indicate no cleavage values for bin Y at position X are present, whereas red cells indicate a large proportion of the cleavage values for that column. (D–F) Mean predicted hydroxyl radical cleavage patterns of CORCS found in (D) the MPSS data set and (E) the Union data set using the discrete sampler, and in (F) the Union data set using the continuous sampler. (G–I) Sequence logos of CORCS found in (G) the MPSS data set and in (H) the Union data set using the discrete sampler, and in (I) the Union data set using the continuous sampler.











