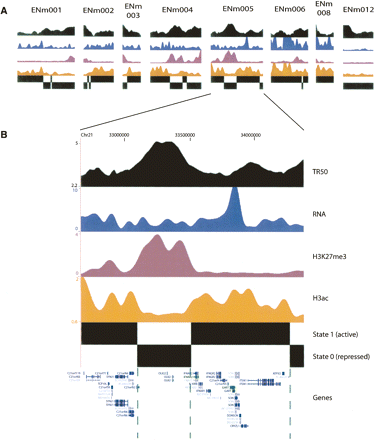
Simultaneous segmentation of four ENCODE functional data types. (A) Exemplary results from eight ENCODE regions ENm001 (1.8 Mb), ENm002 (1 Mb), ENm003 (600 kb), ENm004 (1.7 Mb), ENm005 (1.6 Mkb), ENm006 (1 Mb), ENm008 (1 Mb), and ENm012 (1.2 Mb). For each ENCODE region subpanel, wavelet smoothed data are displayed as tracks ordered top-to-bottom as follows: TR50 (black), RNA (blue), H3K27me3 (purple), and H3ac (orange). State assignments (domains) resulting from simultaneous HMM segmentation are shown at bottom as black rectangles; see Fig. 1 for additional description. (B) Close-up of ENCODE region ENm005, with bracketed intervals indicating exemplary domains in states 0 and 1. State 1 generally corresponds to higher levels of RNA and H3ac and lower levels of TR50 and H3K27me3, and is therefore assigned the active label, while state 0 is correspondingly assigned repressed. The latter contains the oligodendrocyte-specific OLIG1 and OLIG2 genes, which are repressed in the tissues studied under ENCODE.











