Comparison of ChIP-PET-5+ targets and ranked targets from the 50 b every 50-b array ChIP-chip data set
Click on table to view larger version.
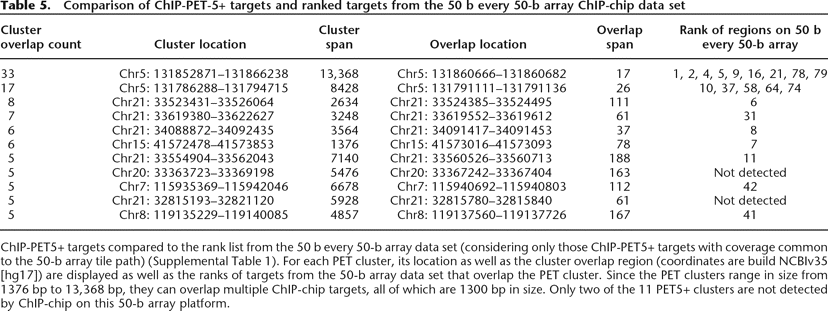
ChIP-PET5+ targets compared to the rank list from the 50 b every 50-b array data set (considering only those ChIP-PET5+ targets with coverage common to the 50-b array tile path) (Supplemental Table 1). For each PET cluster, its location as well as the cluster overlap region (coordinates are build NCBIv35 [hg17]) are displayed as well as the ranks of targets from the 50-b array data set that overlap the PET cluster. Since the PET clusters range in size from 1376 bp to 13,368 bp, they can overlap multiple ChIP-chip targets, all of which are 1300 bp in size. Only two of the 11 PET5+ clusters are not detected by ChIP-chip on this 50-b array platform.











