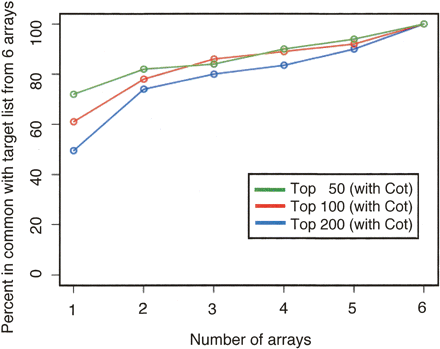
The value of adding biological replicates to a ChIP-chip data set. For the six 50 b every 38-b arrays that were hybridized in the presence of Cot DNA, the reproducibility of target lists for the top 50 (green), 100 (red), and 200 (blue) binding regions was examined as a function of the number of biological replicates analyzed. Each biological replicate is hybridized to a separate array. The agreement is compared against the target list identified by using all six arrays. We see that greater than 80% agreement is obtained when three or more biological replicates are used.











