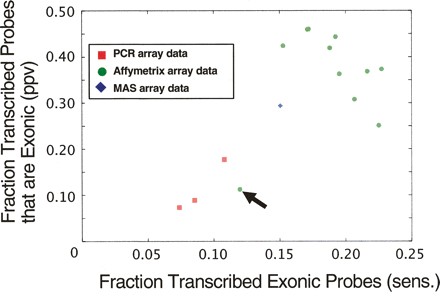
Comparing human chromosome 22 transcription data sets with gene annotation. Transcription data sets were derived from previously published studies. They were generated from three different microarray platforms: PCR (red squares), MAS (blue diamond), and Affymetrix (green circles); in a total of 15 separate experiments (tissues or cell lines), each represented by a point in the figure. We used the RefSeq annotation as a benchmark to assess the quality of the data from each experiment. (x-axis) The fraction of exonic probes that were identified to be transcribed in individual experiments (sensitivity). (y-axis) The fraction of transcribed probes overlapping with an exon (PPV). The PCR tiling-array data were from placenta, fibroblast, and B-cells (Rinn et al. 2003; White et al. 2004), the MAS data from liver (Bertone et al. 2004), and the Affymetrix sets were collected from Kapranov et al. (2002) representing 11 different cell lines. (Arrow) The Affymetrix data from the U87 cell line is not representative since a long section of chromosome 22 is identified as transcriptionally silent, suggesting that this particular experiment probably did not work or something is unusual about U87.











