The peak-finding algorithm ChlPOTle yielded 1008 FAIRE peaks (cutoff = P < 10−25, see Methods)
Click on table to view larger version.
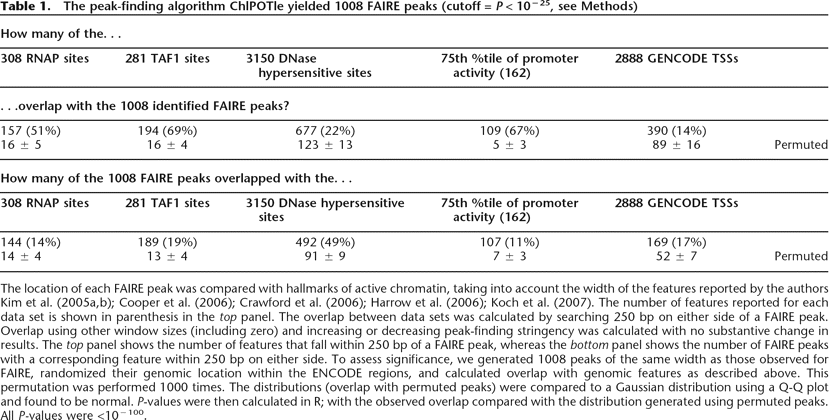
The location of each FAIRE peak was compared with hallmarks of active chromatin, taking into account the width of the features reported by the authors Kim et al. (2005a, b); Cooper et al. (2006); Crawford et al. (2006); Harrow et al. (2006); Koch et al. (2007). The number of features reported for each data set is shown in parenthesis in the top panel. The overlap between data sets was calculated by searching 250 bp on either side of a FAIRE peak. Overlap using other window sizes (including zero) and increasing or decreasing peak-finding stringency was calculated with no substantive change in results. The top panel shows the number of features that fall within 250 bp of a FAIRE peak, whereas the bottom panel shows the number of FAIRE peaks with a corresponding feature within 250 bp on either side. To assess significance, we generated 1008 peaks of the same width as those observed for FAIRE, randomized their genomic location within the ENCODE regions, and calculated overlap with genomic features as described above. This permutation was performed 1000 times. The distributions (overlap with permuted peaks) were compared to a Gaussian distribution using a Q-Q plot and found to be normal. P-values were then calculated in R; with the observed overlap compared with the distribution generated using permuted peaks. All P-values were <10−100.











