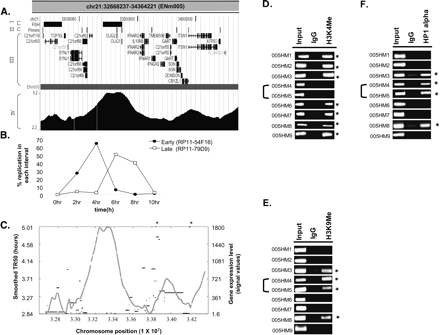
Replication profile demarcates chromosomal domains. (A) UCSC Genome Browser display of a 1.7-Mb region from Chromosome 21 (ENm005). This Browser picture highlights four tracks (I–IV): (I) FISH: chromosomal location of BAC clones (RP11-54F16 and RP11-79D9, from left to right) selected for the interphase FISH experiment shown in B; (II) Primers: chromosomal locations of the primers (005HM1–9, left to right) selected for ChIP assay to ascertain the histone modifications and HP1α-binding sites shown in D–F; (III) RefSeq: positions of all the genes in this chromosomal segment; and (IV) the contiguous TR50 profile. (B) Dual color FISH was performed with HeLa cells synchronously progressing through S phase. RP11-54F16 (from early-replicating area on left) was labeled with spectrum red dUTP, while RP11-79D9 (from late-replicating area) was labeled with spectrum green dUTP. Dual color FISH with these two BAC clones ascertained the replication time of the two regions of Chromosome 21 set 355 kb apart. (C) Plot of smoothed TR50 (Y-axis on left, gray) against level of transcription of genes (Y-axis on right, black). The two asterisk marks represent transcripts whose transcription levels exceeded the Y-axis limit (i.e., 2346 and 10,010 for left and right asterisks, respectively). (D–F) ChIP-PCR assay across ENm005 region (see Supplemental Table 3 for primers). PCR was performed on DNA chromatin immunoprecipitated with antibodies against methylated histones (H3 Lys4 and H3 Lys9) and HP1α. (Input) DNA control before immunoprecipitation; (IgG) ChIP with rabbit IgG was negative control for nonspecific precipitation. Forty cycles of PCR were performed for H3 Lys4 and HP1α and 30 cycles for H3 K9 di-Me. The asterisks refer to primer pairs that gave positive ChIP signal for the indicated antibodies relative to the IgG negative control. 005HM4 and 005HM5 were from the late-replicating island in ENm005.











