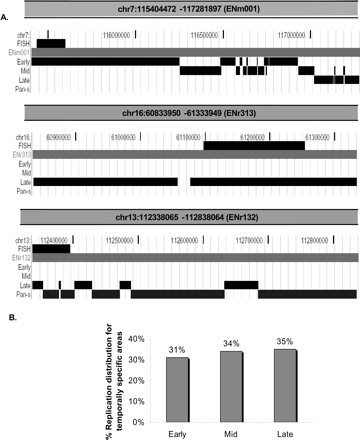
Segregation of chromosomal segments with temporally specific and temporally nonspecific pattern of replication. (A) On the basis of TR50, temporally specific regions are further segregated and displayed as three tracks (UCSC Genome Browser), early-, mid-, or late-replicating, while chromosomal regions undergoing temporally nonspecific replication are highlighted under the pan-S track. The three panels in this figure show segregation of replication timing for three chromosomal segments. (Top panel) The 1.9-Mb region (ENm001) of Chromosome 7; (middle and bottom panels) examples of two 500-kb chromosomal segments from Chromosomes 16 (ENr313) and 13 (ENr132) that underwent late and pan-S replication, respectively. The FISH track in all the three panels refers to the chromosomal positions of BAC clones selected for the interphase FISH experiment shown in Figures 3 and 4. (B) Percent of temporally specific chromosomal segments replicating in early-, middle-, or late-S phase.











