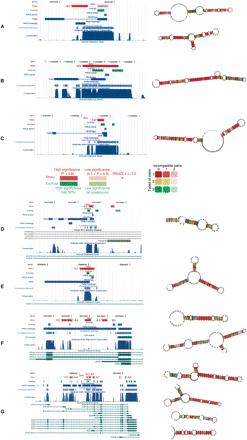
Selected high scoring examples. (Left) UCSC Genome Browser screenshots featuring conserved RNA predictions and additional ENCODE analysis tracks are shown. The significance levels of RNAz and EvoFold hits are color coded (see legend). (*) Significant AlifoldZ hits; the Z-score is shown. In addition, the results of the RACE/microarray experiments, TARs/Transfrags, constrained elements, phastCons scores, and GENCODE annotations are shown. For details on these tracks, refer to Methods. (Right) Consensus structure models generated by RNAalifold are shown for selected hits (marked by gray, dashed boxes; in example G, the first three hits and the sixth hit are shown). In the consensus structures, variable positions are circled indicating compensatory and consistent mutations supporting the structure. The color indicates the number of different nucleotide combinations forming one base pair. Inconsistent mutations lead to pale colors. Examples A–C show predicted structures in intergenic regions. Examples D and E are located in introns of protein-coding regions. Examples F and G show structures associated with alternative spliced transcripts of protein-coding loci detected by the GENCODE project. For further information, refer to the text.











