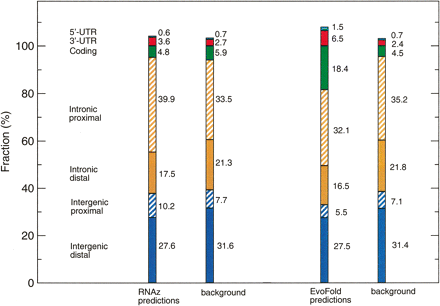
Genomic location of predicted RNAs (high significance level) relative to the GENCODE protein gene annotation. For comparison, the annotation of the input alignments is shown for both RNAz and EvoFold (they differ slightly because of the different filtering steps used for each program; see Methods). “Distal” and “Proximal” refer to a distance boundary of 5 kb away from the next gene (intergenic fraction) or coding exon (intronic fraction). Some hits fall within more than one annotation category, thus the sums of the fractions are slightly >100%.











