A comparison of over-represented motifs in OCT4 target promoters using different bioinformatics approaches
Click on table to view larger version.
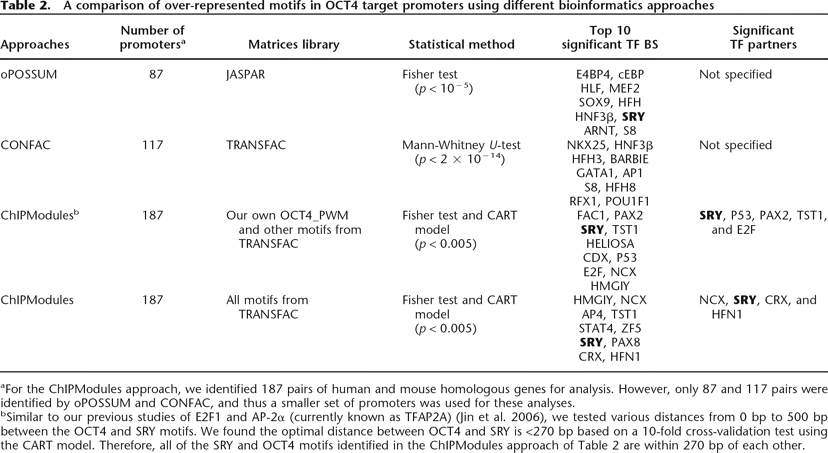
aFor the ChIPModules approach, we identified 187 pairs of human and mouse homologous genes for analysis. However, only 87 and 117 pairs were identified by oPOSSUM and CONFAC, and thus a smaller set of promoters was used for these analyses.
bSimilar to our previous studies of E2F1 and AP-2α (currently known as TFAP2A) (Jin et al. 2006), we tested various distances from 0 bp to 500 bp between the OCT4 and SRY motifs. We found the optimal distance between OCT4 and SRY is <270 bp based on a 10-fold cross-validation test using the CART model. Therefore, all of the SRY and OCT4 motifs identified in the ChIPModules approach of Table 2 are within 270 bp of each other.











