Table 1.
Classification estimates of Sn and Sp rates for OCT4 target promoters in Ntera2 cells
Click on table to view larger version.
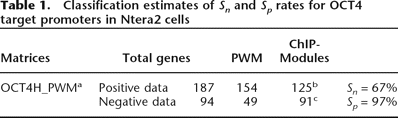
aThe OCT4H_PWM was built from our ChIPMotifs approach using an optimal cutoff threshold of 0.88 for Sc_h and 0.85 for Sp_h to identify the OCT4-binding sites.
bThe number of promoters that contained one of the five identified modules.
cThe number of promoters that lacked one of the five identified modules.











