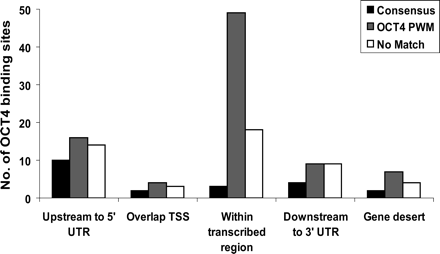
A comparison of the distribution of binding sites that (1) contain a conventional OCT consensus (ATGC[A/T]AAT); (2) lack the consensus but contain a match to the OCT PWM identified by the ChIPMotifs approach with 1.0 > Sc_h ≥ 0.88 and 0.95 > Sp_h ≥ 0.85 (the consensus sites would have also been identified by the OCT PWM but have not been included in this set for comparison purposes); and (3) contain no match to the OCT PWM above Sc_h 0.88, Sp_h 0.85. The OCT4-binding regions were defined as peaks detected on duplicate ENCODE arrays using a peak calling program developed for ChIP-chip experiments (Bieda et al. 2006); the gene list was based on GENCODE Genes (Harrow et al. 2006).











