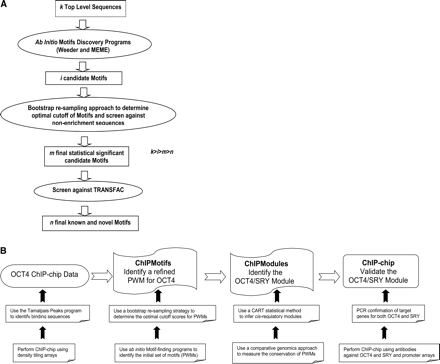
(A) A diagram of the integrated ChIPMotifs approach. Beginning with a set of k high-confidence level ChIP-chip sequences, ab initio motifs discovery programs such as Weeder and MEME are used to identify i candidate motifs. Then, a bootstrap resampling approach is used to determine cutoffs for the motifs and to screen against nonenriched sequences to obtain m final statistically significant candidate motifs. Finally, the motifs are screened against TRANSFAC to retrieve n known and novel motifs. (B) A strategy diagram showing how our ChIPMotifs and ChIPModules approaches work in concert to efficiently mine OCT4 ChIP-chip data, to allow the development of de novo OCT4 motifs, to identify new cis-regulatory modules of OCT4 and SRY, and finally to develop an experimentally confirmed set of OCT4 and SRY targets.











