Enrichment of motifs in ChIP hits
Click on table to view larger version.
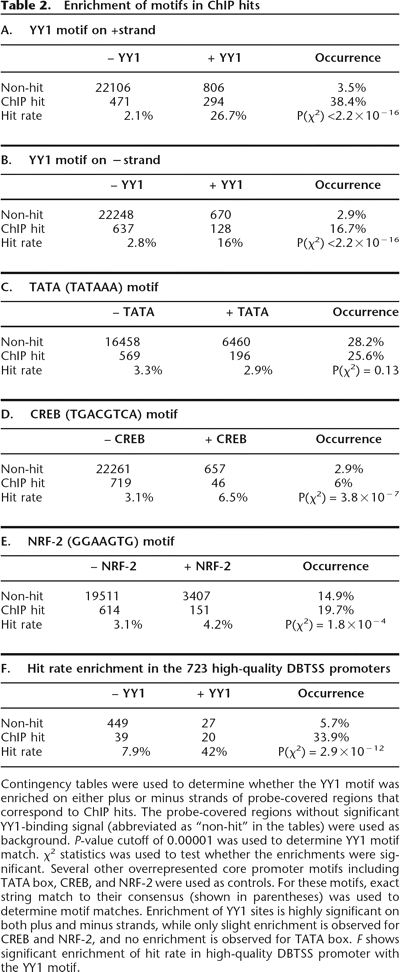
Contingency tables were used to determine whether the YY1 motif was enriched on either plus or minus strands of probe-covered regions that correspond to ChIP hits. The probe-covered regions without significant YY1-binding signal (abbreviated as “non-hit” in the tables) were used as background. P-value cutoff of 0.00001 was used to determine YY1 motif match. χ2 statistics was used to test whether the enrichments were significant. Several other overrepresented core promoter motifs including TATA box, CREB, and NRF-2 were used as controls. For these motifs, exact string match to their consensus (shown in parentheses) was used to determine motif matches. Enrichment of YY1 sites is highly significant on both plus and minus strands, while only slight enrichment is observed for CREB and NRF-2, and no enrichment is observed for TATA box. F shows significant enrichment of hit rate in high-quality DBTSS promoter with the YY1 motif.











