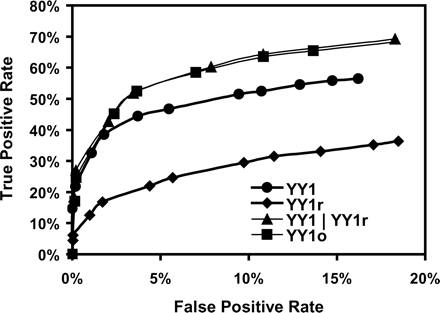
The YY1 PSSM accurately predicts ChIP hits. The YY1 PSSM was searched against probe-covered regions of the 765 ChIP hits (true positives) and the 10,000 probe-covered regions with the lowest ChIP signal (true negatives). True positive rates were plotted against the false positive rates at various P-value cutoffs. (YY1) YY1 motif on the plus strand; (YY1r) YY1 motif on the minus strand; (YY1/YY1r) YY1 motif on either the plus or the minus strand; (YY1o) YY1 motif after optimization in ROVER algorithm on either the plus or the minus strand.











