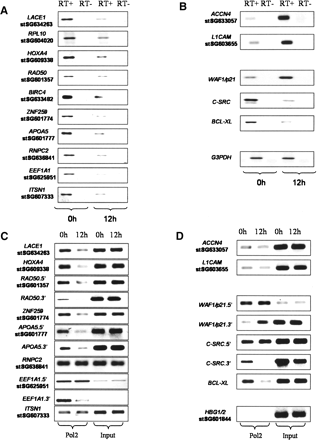
Histone deacetylation at promoter regions results in decreased expression of the associated genes mediated at the transcriptional level. RNAs were prepared from cells that were either untreated (0 h) or treated with 5 mM Na-butyrate for 12 h. Total RNA (1 μg) was used for preparing gene-specific cDNAs using reverse transcriptase (RT), which were analyzed by RT–PCR to investigate the RNA levels of genes displaying histone deacetylation (A), while genes with increased acetylation, genes from the literature, and a loading control (GAPDH) are presented in B. Similar results were obtained in a biological replicate or when cDNAs were obtained using oligo(dT). ChIPs were prepared from cells that were either nontreated (0 h) or treated with 5 mM Na-butyrate for 12 h using an antibody against the C-terminal domain of POLR2A. The analyzed regions included most from Supplemental Table 1, divided into those with histone deacetylation (C) and with histone hyperacetylation from previous literature or serving as negative control (HBG1/2) (D). For some genes, two different genomic regions were analyzed, one close to the TSS (Supplemental Table 1) is indicated by the gene name, followed by 5′, while an intragenic region was analyzed using the same primers used in A and B and is indicated by the gene name, followed by 3′. Similar results were obtained in two other biological replicates.











