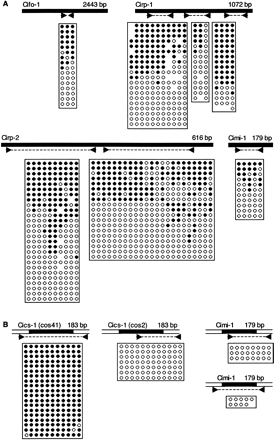
Variable DNA methylation of repetitive elements in C. intestinalis matches flanking DNA. Elements as named here or by Simmen and Bird (2000) are diagrammed as solid bars above each bisulfite profile showing primer pair location. A horizontal row within each box corresponds to one sequenced clone in which specific CpGs were methylated (solid) or unmethylated (open). Mutated CpG sites are shown as blanks within a row. (A) Results using primer sets within the element showed a mixture of methylated and unmethylated clones. (B) Locus-specific primers showed that the methylation patterns of two Cics-1 elements and two Cimi-1 elements at specific genomic locations are homogeneous and match the methylation status of flanking genomic DNA (open bars above). The locations of the elements analyzed in B are shown in Fig. 1.











