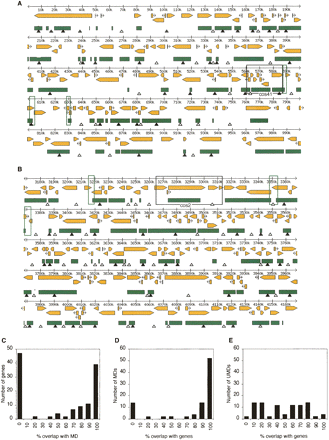
Methylated domains (MDs) within two 1-Mb genomic regions from chromosome 7q (A) and chromosome 4q (B) colocalize with transcription units. MDs (green bars) were inferred from CpG[o/e] ratios as described in the text. The inferred pattern was experimentally verified by methylation-sensitive PCR at sites marked by closed triangles (methylated) and open triangles (unmethylated). Yellow arrows show the size and orientation of predicted transcription units. The positions of bisulfite sequenced cosmid inserts cos41 and cos2 (see Fig. 1) are shown by black boxes. Green boxes indicate regions that were bisulfite sequenced in order to test the accuracy of predicted unmethylated domain (UMD)/MD boundaries (Supplemental Fig. 2). (C) Frequency of predicted genes that overlap to varying degrees with MDs. (D) Frequency of MDs that overlap to varying degrees with predicted genes. (E) Frequency of UMDs with gene overlap.











