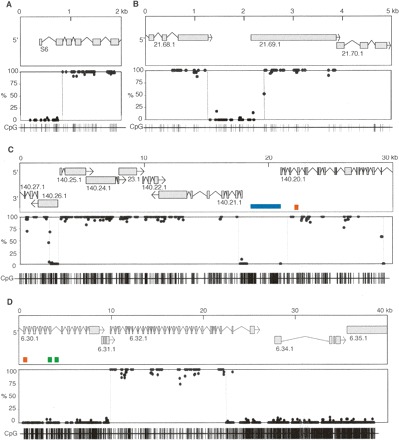
A mosaic pattern of methlyated domains (MDs) and unmethylated domains (UMDs) in the C. intestinalis genome. Bisulfite sequencing showed the DNA methylation status at every CpG site in ribosomal protein S6 (scaffold 13: 227234–231211) (A), a randomly selected region of chromosome 9q (2421536–2429959) (B), a region of chromosome 7q corresponding to 29,300 bp of cosmid insert cos41 (C), and a region of chromosome 4q corresponding to 38,500 bp of cosmid insert cos2 (D). The top panels show gene annotation and plots of CpG methylation are shown below. Each dot represents a specific CpG corresponding to the CpG map at the bottom of each figure. MD/UMD boundaries are shown by broken lines. Repetitive sequences Cifo-1 (blue), Cimi-1 (green), and Cics-1 (orange) are shown.











