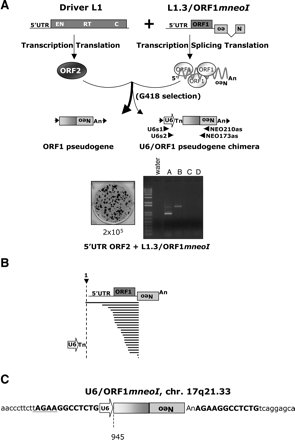
Template choice and template switching are not mutually exclusive. (A) Rationale and results of the assay. The trans-complementation assay is described in Figure 2. Retrotransposition assays were conducted by transfecting 2 × 105 HeLa cells with the driver (5′ UTR ORF2 No neo) and reporter (L1.3/ORF1mneoI) plasmids. Genomic DNAs from pools of G418-resistant foci were subjected to individual nested PCR reactions using the indicated primers (U6s1-NEO210as and U6s2-NEO173as) to detect U6/ORF1mneoI pseudogenes. (B) Results of the assay. Sequence analysis of 23 PCR products confirmed the existence of the U6/ORF1mneoI pseudogenes. A full-length ORF1mneoI mRNA is represented at top. The U6 sequence ends in four to six thymidine residues and is followed by ORF1mneoI RNA. The horizontal bold lines indicated the amount of the L1 included in the pseudogene. The longest U6/ORF1mneoI extends beyond the first nucleotide of ORF1mneoI RNA, indicating that the transcript was initiated from the CMV promoter upstream of the reporter gene. (C) The structure of a U6/ORF1mneoI pseudogene. The characterization of a single U6/ORF1mneoI pseudogene by inverse PCR revealed the hallmarks of L1 retrotransposition. Top bold characters on each side of the pseudogene represent the target site duplication. Lowercase characters represent the flanking genomic sequence. The complement of the L1 endonuclease cleavage recognition site (5′-TTCT/A) is underlined. The numbering is based on the L1.2 reference sequence (Dombroski et al. 1991).











