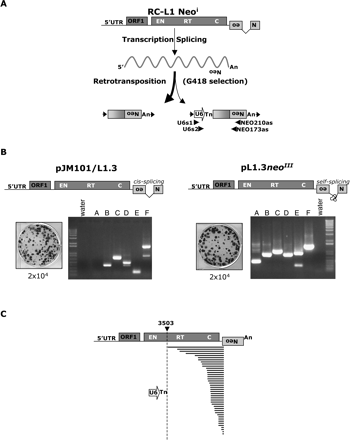
A cultured cell assay to detect U6/L1 pseudogenes. (A) Rationale of the assay. The 3′ UTR of a retrotransposition-competent L1 (RC-L1) was tagged with a retrotransposition indicator cassette (light gray box labeled with a backward Neo). ORF1 and ORF2 are indicated by the dark-gray rectangles and the relative positions of the endonuclease (EN), reverse transcriptase (RT), and cysteine-rich domains (C) of ORF2 are indicated. Cartoons depicting the structures of the resultant retrotransposition events that confer G418 resistance (G418R) to HeLa cells are shown at the bottom of the diagram. A 5′-truncated L1 insertion is shown at the left and a U6/L1 pseudogene is shown at the right. Target site duplications flanking the elements are represented as horizontal arrows. PCR primers used to detect U6/L1 pseudogenes are indicated below the U6/L1 schematic. (B) Results of the assay: Shown are schematic representations of two RC-L1s (pJM101/L1.3 and pL1.3neoIII). pJM101/L1.3 contains our standard retrotransposition indicator cassette (Moran et al. 1996). pL1.3neoIII contains a retrotransposition indicator cassette disrupted by a self-splicing group I intron (Esnault et al. 2002; Dewannieux et al. 2003). Retrotransposition assays conducted by transfecting 2 × 104 HeLa cells revealed that both L1s retrotranspose at similar efficiencies (Wei et al. 2000). Genomic DNAs derived from six independent pools of G418-resistant foci then were used as templates in nested PCR reactions with the primers noted in A to detect the U6/L1 pseudogenes (lanes A–F). Molecular size standards are indicated on the gels (1-kb ladder plus from Invitrogen). (Water) PCR reactions conducted without the genomic DNA template. (C) Structures of the U6/L1 pseudogenes. Sequence analysis of the PCR products amplified in B confirmed the existence of the U6/L1 pseudogenes. All sequences contain the 3′ terminus of U6 (depicted as an horizontal open arrow) and a variable 5′−truncated L1. A full-length RC-L1 with the spliced reporter cassette is represented at the top. The U6 sequence ends in 4–8 thymidine residues and is followed by a 5′-truncated L1. The horizontal bold lines indicate the amount of the L1 sequence included in the pseudogene. The longest sequence extends to nucleotide 3503 of the L1. The numbering is based on the L1.2 reference sequence (Dombroski et al. 1991).











