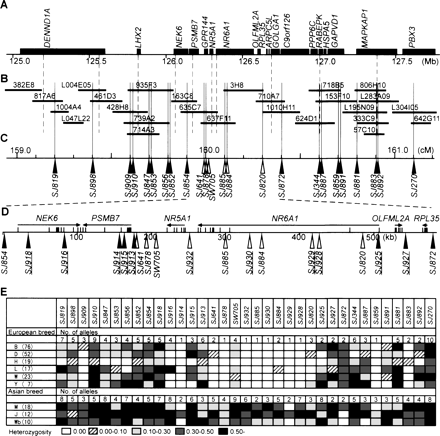
Analyses of genomic structure and genetic variation of the QTL region. (A) Gene map of a part of the human chromosome 9qter. (B) BAC contig for the QTL region. Vertical lines indicate positions of STS (broken) and microsatellite markers (solid). BAC clones (Suzuki et al. 2000) from which markers were developed and those in the minimum tiling path are shown. (C) Linkage map of microsatellite markers on SSC1qter. Arrowheads indicate positions of microsatellite markers, and clear arrowheads indicate those lacking genetic variation in European commercial breed pigs. (D) Defining the QTL region by the genetic variation of microsatellite markers. Sequencing analysis was performed for the region from SJ854 to SJ872. Exons of the genes found in this region are indicated, and arrows show the directions of gene transcription. We isolated 11 novel markers (underlined) and added them to the analysis of genetic variation. A reduction of genetic variation of microsatellite markers in European commercial breed pigs occurred between SJ641 and SJ820. The GenBank accession number for the genomic sequence in this region is AP009124. (E) Heterozygosities of markers in each breed. Breeds are: B, Berkshire; D, Duroc; H, Hampshire; L, Landrace; W, Large White; Y, Yorkshire; M, Meishan; J, Jinhua; and Wb, Japanese wild boar; and numbers of samples are shown in parentheses.











