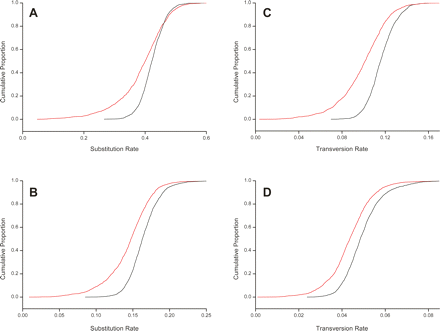
Nucleotide substitution and transversion rates are suppressed within macroRNA transcripts. Panels show the cumulative distributions of substitution (A,B) and transversion rates (C,D) as measured on macroRNA transcripts (red curves), and the same rates measured on nearby nonoverlapping AR sequence of matched length (black curves). (A) Mouse–human substitution rates; (B) mouse–rat substitution rates; (C,D) mouse–human and mouse–rat transversion rates. All macroRNA rates are significantly different from, and lower than, the putatively neutral AR rates (Kolmogorov-Smirnov test, P < 10−15 for all panels). The suppression of transversion rates in macroRNAs compared to AR sequence demonstrates that these observations are not a consequence of a higher density of highly mutable CpG sites within ARs, since the associated mutations are mainly transitions rather than transversions.











