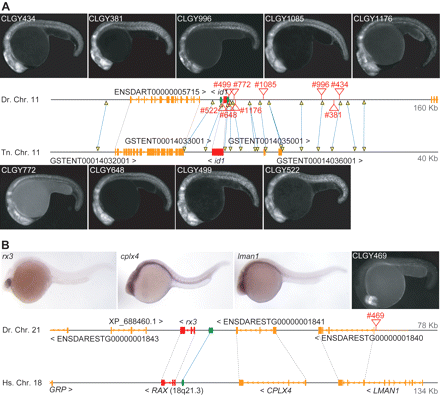
Genomic regulatory blocks are extended regions of cis-regulatory content. (A) Zebrafish id6 locus is orthologous to tetraodon id1, located in a gene desert spanned by conserved noncoding elements (yellow triangles connected with blue lines, corresponding to Ensembl translated BLAT alignments). Two HCNEs are conserved from fish to human id1 loci (green ovals in zebrafish locus). The insertions recovered are labeled as red triangles with transgenic line numbers. Note the same global expression pattern regardless of insertion position (id6 expression pattern in Supplemental Fig. S1). (B) GRB of human RAX and zebrafish rx3 loci, consisting of RAX, CPLX4, and LMAN1 genes. An insertion within lman1, CLGY469, assumes the expression pattern of rx3, not of the gene it is inserted into. While only one HCNE can be discerned in this alignment, multiple elements are found in fish/fish alignments (data not shown).











