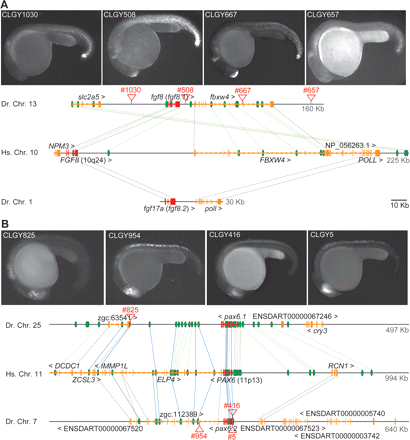
(A) The duplicated fgf8 loci of zebrafish. (Upper track) The current fgf8 locus has kept its downstream neighbor fbxw4 but has lost the farther downstream NP_056263.1 and POLL genes, which are, however, retained downstream from fgf17a. NP_056263 is conserved in zebrafish at this position, but currently not annotated (data not shown). Both duplicated loci have lost neighboring genes, even though the syntenic relationship of human NPM3, FGF8, and FBXW4 is conserved across chordate genomes. HCNEs located in the upstream gene slc2a5 are found far downstream from FGF8 in human, with no evidence of a human counterpart of slc2a5. Numerous HCNEs were also found within FGF8, downstream from the gene, and inside the bystander FBXW4, and most of them are conserved only in zebrafish fgf8. Enhancer detection insertions (red triangles) cover the entire GRB and show partial (CLGY1030) or complete fgf8 expression patterns at 24 h. In these and all other images, anterior is to the left and dorsal to the top. (B) Human PAX6 GRB covers 1 Mb, containing five bystander genes. One of these, the far downstream DCDC1 is not found in teleost genomes; of the others, only one copy has been retained either in pax6.1 or pax6.2 GRBs. Insertion CLGY825 in the pax6.1 GRB shows the correct expression pattern (cf. Supplemental Fig. S1) despite being adjacent to the ZCSL3 ortholog, while CLGY954 in the pax6.2 GRB is inside elp4 yet has the expression pattern of pax6.2 (cf. CLGY5 and −416 inside pax6.2 and pax6.2 expression pattern in Supplemental Fig. S1).











