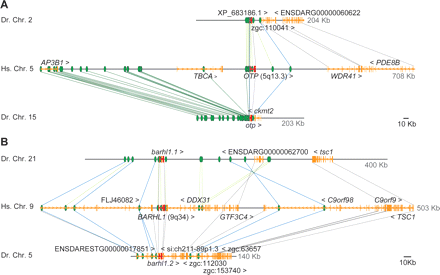
Duplicated zebrafish GRBs. (A) Orthopedia (otp). (Green ovals) HCNEs; (orange and red gene models) bystander and target genes, respectively. After duplication, one of the zebrafish GRBs (upper track) lost most of its upstream parts, including a large HCNE array and AP3B1 and TBCA genes, and AP3B1 and TBCA landed elsewhere in the genome (data not shown), while the other (lower track) kept most of the HCNE array while AP3B1 and TBCA were lost by neutral evolution. Cross-species sequence comparisons indicate that the region upstream of ckmt2 harbors an RASGRF2 ortholog (data not shown) for which no full-length cDNA is available. (B) The duplicated barhl1 loci. Barhl1.1 (upper track) lost the bystander genes DDX31, C9orf98, and C9orf9, but retained all of the HCNEs found in the human locus, and there is evidence of an inversion that occurred between barhl1.1 and tsc1 (crossed lines interconnecting HCNEs), which includes the only retained copy of the GTF3C4 gene. Barhl1.2 (lower track) lost the downstream GTF3C4, but retained all other annotated genes and some HCNEs.











