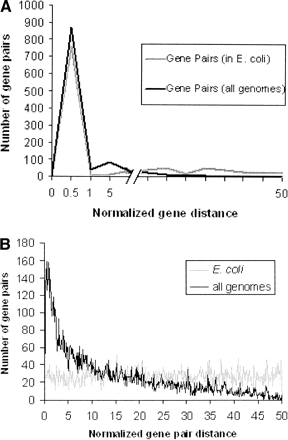
Normalized gene distance distributions among the data sets used for prediction of protein–protein interactions. The normalized gene pair distance is defined as the number of base pairs that separate the translation start sites of two genes in a pair, divided by the total genome length and then scaled to 100. (A) (Gray) Normalized distances between the known interacting protein pairs in E. coli; (black) the minimum normalized distance between the same proteins on any of the other genomes. (B) Normalized distances between the protein pairs, which are hypothesized to be noninteracting. (Gray line) The normalized distance distribution in E. coli; (black line) the minimum of this between the same pair of proteins in any other genome.











