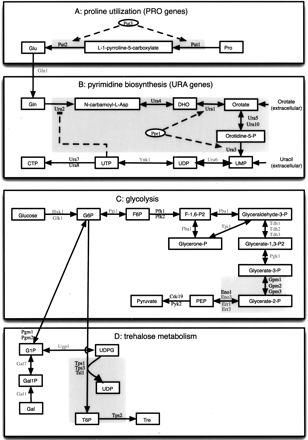
The metabolic pathways affected in the gene deletion mutants used in this study. (Bold) Deleted genes, (shaded) metabolic co-sets, identified using flux coupling analysis (see text). (A) Proline utilization (PUT genes). (B) Pyrimidine biosynthesis (URA genes). (DHO) Dihydroorotate. (C) Glycolysis. (G6P) Glucose 6-phosphate, (F6P) fructose 6-phosphate, (F-1,6-P2) fructose-1,6-bisphosphate, (Glycerone-P) dihydroxyacetone phosphate, (Glyceraldehyde-3-P) glyceraldehyde 3-phosphate, (Glycerate-1,3-P2) 1,3-bisphosphoglycerate, (Glycerate-3-P) 3-phosphoglycerate, (Glycerate-2-P) 2-phosphoglycerate, (PEP) phosphoenolpyruvate. (D) Trehalose synthesis. (G1P) glucose 1-phosphate, (Gal1P) galactose 1-phosphate, (Gal) galactose, (Tre) trehalose, (T6P) trehalose-6-phosphate.











