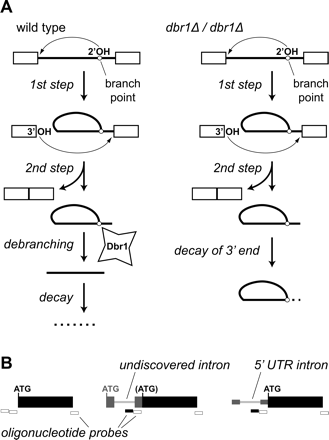
Sequences that accumulate in a debranchase mutant are used to identify new introns. (A) Intron removal occurs via two trans-esterification reactions (thin arrows), carried out by the 2′- and 3′-hydroxyls indicated. The resulting intron lariats are linearized by a debranching enzyme (Dbr1) and degraded by exonucleases (left). In a debranchase mutant (right), intron sequences upstream of the branch point are inaccessible to exonucleases, and accumulate to high levels. (B) When dbr1 mutant cDNA is compared to wild-type cDNA on our microarrays, probes specific to the region between the 5′-splice site and the branch point of an expressed intron show an intensity bias (black probes), while most other probes do not (white probes). (Boxes) Exons; (lines) introns; (ATG) translation start codons; (gray items) previously undiscovered introns and exons.











