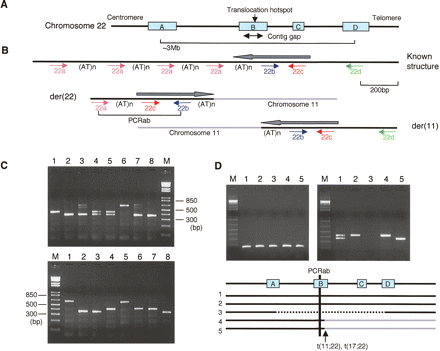
Development of a PCR system specific for the PATRR22 region. (A) LCR22 in 22q11.2. There are at least four copies of LCR22, LCR22A, LCR22B, LCR22C, and LCR22D. One of the LCR22s, LCR22B, is the site of a genomic contig gap as well as the translocation breakpoint hotspot. (B) Location and orientation of the four PCR primers on the known structure appearing in other LCR22s and the t(11;22) translocation derivative chromosomes. There are multiple copies of primer “a” as a result of tandem repeats. Black lines indicate chromosome 22, while light-gray lines indicate chromosome 11. All of the genomic clones present in the database manifest the “known structure.” Only a der(22)-containing clone should be positive for the PCRab products. “(AT)n”s indicate the AT-rich region. Gray arrows indicate the putative PATRR22 arms. (C) Representative results of the PCRab reaction on genomic DNA. (Top) PCRab for eight normal individuals; (bottom) PCRab for patients with the 22q11.2 deletion syndrome. The additional bands originating from heteroduplexes appear as PCR artifacts in lanes 3 and 7. (D) The PCRbc (left) and PCRab (right) for somatic cell hybrids. The positive PCRbc product arises as a result of the presence of any of the LCR22s in the hybrids, while the positive PCRab product is derived only from LCR22B. (Lane 1) Normal human; (lane 2) GM10888, a human/hamster hybrid containing a normal chromosome 22 as its only intact human chromosome; (lane 3) Cl6-2/EG, a human/hamster hybrid containing a del(22q11.2) chromosome; (lane 4) Cl-4/GB, a human/hamster hybrid cell line containing the der(22) of a constitutional t(11;22); (lane 5) GM11685 (NF13/D3), a subclone of a human/mouse hybrid cell line containing the der(22) of a constitutional t(17;22). Black solid lines in the diagram below represent the chromosome 22q11 regions present in the hybrid lines analyzed.











