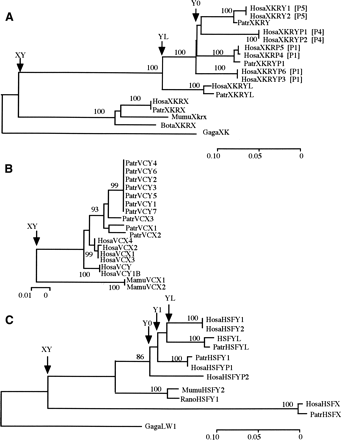
NJ trees of (A) XKRY, (B) VCY, and (C) HSFY. Whenever chicken homologs are available, they are used for rooting the trees. In A, palindromic locations of human XKRY copies are indicated by bracketed P1, P4, and P5. The numbers next to nodes show the bootstrap values in 1000 replications. Node XY, YL, and Y0 stand for X/Y differentiation, retroposition from Y to autosome or vice versa, and coalescence of all human Y copies, respectively. The accession numbers of the DNA sequences and the sequence alignments are given in Supplemental Table 1 and Supplemental Figure 1, respectively. The gene nomenclature follows a four-letter prefix system to indicate genus and species names: (Hosa) Homo sapiens; (Patr) Pan troglodytes; (Mamu) Macaca mulatta; (Mafa) Macaca fascicularis; (Paha) Papio hamadryas; (Ceap) Cebus apella; (Saoe) Saguinus oedipus; (Sasc) Saimiri sciurea; (Cafa) Canis familiaris; (Susc) Sus scrofa; (Bota) Bos taurus; (Mumu) Mus musculus; (Rano) Rattus norvegicus; (Maeu) Macropus eugenii; (Smma) Sminthopsis macroura; (Gaga) Gallus gallus.











