Rice homologs in Poaceae, non-Poaceae monocots, and other plant species
Click on table to view larger version.
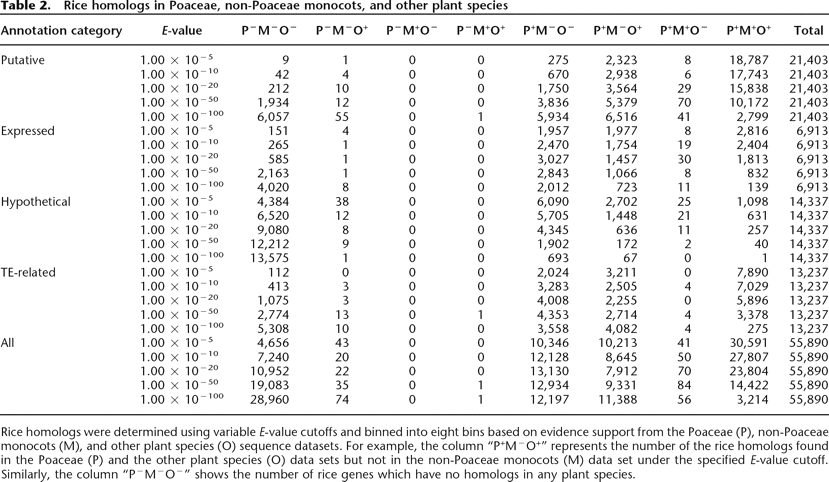
Rice homologs were determined using variable E-value cutoffs and binned into eight bins based on evidence support from the Poaceae (P), non-Poaceae monocots (M), and other plant species (O) sequence datasets. For example, the column “P+M−O+” represents the number of the rice homologs found in the Poaceae (P) and the other plant species (O) data sets but not in the non-Poaceae monocots (M) data set under the specified E-value cutoff. Similarly, the column “P−M−O−” shows the number of rice genes which have no homologs in any plant species.











