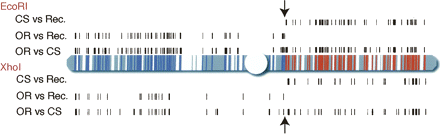
Recombination breakpoint mapping using RAD markers in Drosophila. Polymorphic RAD markers between Oregon-R (OR) and Canton-S (CS) flies were identified on a genomic tiling path microarray. Black tick marks above the diagrammed second chromosome represent the genomic locations of EcoRI RAD tags with an average differential hybridization greater than twofold. Tick marks below the chromosome represent XhoI RAD tags with a differential hybridization >2.3-fold. Polymorphic markers are found when the recombinant line (Rec.) is compared to the OR parental line on the left arm of the chromosome and when compared to the CS parental line on most of the right arm of the chromosome. Marker patterns indicative of CS inheritance are shown as blue regions of the chromosome, and OR inheritance as red. Arrows mark the recombination breakpoint. Large gaps in the tiling array are shown as white regions of the chromosome.











