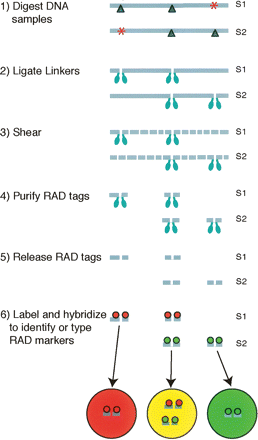
Restriction site associated DNA (RAD) markers can be identified and typed by detecting differential hybridization patterns of RAD tags on a microarray. Genomic DNA samples S1 and S2 contain the recognition sequence for various restriction enzymes at locations throughout the genome. Dark blue triangles represent restriction sites of a particular enzyme. Some of these restriction sites are only present in one sample because of polymorphisms that disrupt the recognition sequence (red asterisks). The two samples are separately digested with a particular restriction enzyme and then ligated to biotinylated linkers (light blue ellipses). The DNA is randomly sheared leaving only the fragments that were directly flanking a restriction site attached to biotin linkers. These fragments are purified using streptavidin beads and released by digestion at the original restriction site. Loci containing polymorphisms, such as the left locus of S2 or the right locus of S1, will not contain tags for that locus in the purified RAD-tag sample, thus resulting in differential hybridization patterns of RAD tags on a microarray.











