Table 1.
List of predicted inversions for which there is some form of evidence supporting the inverted orientation
Click on table to view larger version.
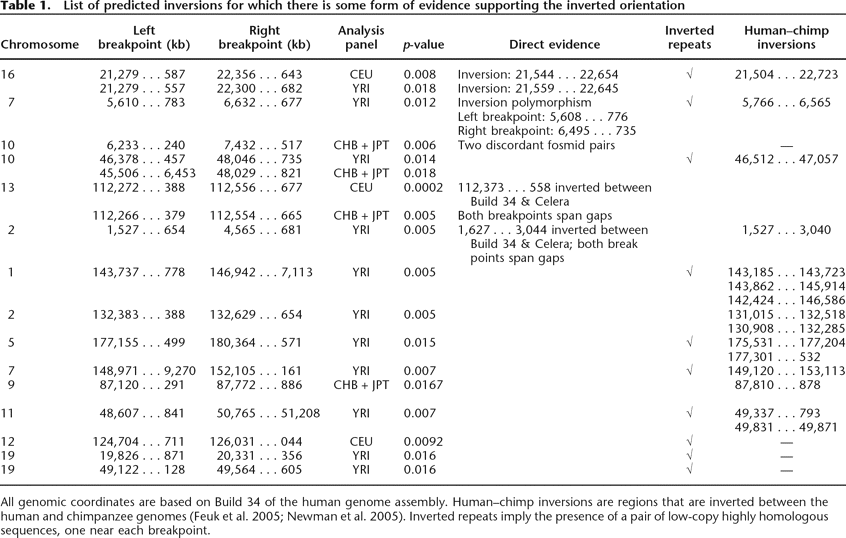
All genomic coordinates are based on Build 34 of the human genome assembly. Human–chimp inversions are regions that are inverted between the human and chimpanzee genomes (Feuk et al. 2005; Newman et al. 2005). Inverted repeats imply the presence of a pair of low-copy highly homologous sequences, one near each breakpoint.











