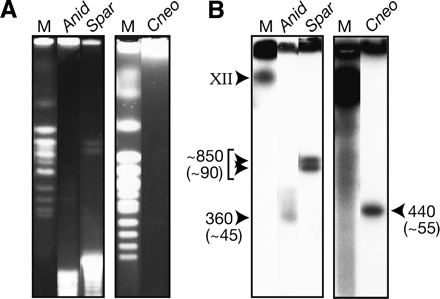
Determination of rDNA array sizes by pulsed field gel electrophoresis. (A) Ethidium bromide-stained gels showing the sizes of the rDNA arrays from A. nidulans, S. paradoxus, and C. neoformans after digestion of chromosomal plugs with HinDIII, BamHI, and AgeI, respectively. (B) Southern blot of the gels from A probed with a conserved region of the LSU from S. cerevisiae to confirm the rDNA bands. Array sizes (kilobase) are indicated, as are rDNA copy numbers (in parentheses), and these were calculated using S. cerevisiae chromosomes as size markers (M).











