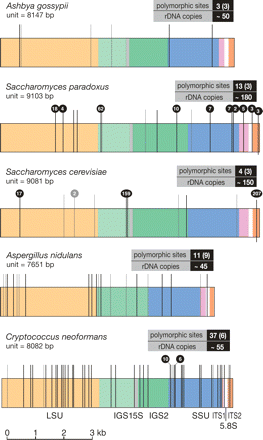
Positions of the polymorphisms in the rDNA unit. Polymorphic sites for each species are plotted onto a map of a single rDNA unit. rDNA features are color coded as indicated beneath the bottom rDNA unit. High-confidence polymorphisms are shown as black lines, low-confidence polymorphisms as gray lines, and the total numbers are boxed (low-confidence polymorphisms in parentheses). Although the polymorphisms are shown in a single repeat, in reality they are likely to be scattered throughout the array. Polymorphisms present in more than one sequence read are indicated by black balls above the line, with the number of reads shown in the ball. In many cases these are likely to result from the coverage level of the whole-genome shotgun sequence data. The rDNA unit length and copy number (from Fig. 3) are also indicated (S. paradoxus is the diploid copy number). Diagram is to scale.











